

Probability-based protein identification by searching sequence databases using mass spectrometry data.
De novo protein sequence analysis software#
PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. A common open representation of mass spectrometry data and its application to proteomics research. Mass spectrometry data analysis in the proteomics era. Informatics for protein identification by mass spectrometry. Computational analysis of shotgun proteomics data. Large-scale protein identification using mass spectrometry. Electron capture dissociation for structural characterization of multiply charged protein cations. Electron-capture dissociation tandem mass spectrometry. New prospects for proteomics - electron-capture (ECD) and electron-transfer dissociation (ETD) fragmentation techniques and combined fractional diagonal chromatography (COFRADIC). Protein sequencing by mass analysis of polypeptide ladders after controlled protein hydrolysis. Prediction of low-energy collision-induced dissociation spectra of peptides. Mobile and localized protons: a framework for understanding peptide dissociation. Statistical characterization of ion trap tandem mass spectra from doubly charged tryptic peptides. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Fragmentation pathways of protonated peptides. Towards understanding the tandem mass spectra of protonated oligopeptides. Interpreting mass-spectra of multiply charged ions. Statistical characterization of the charge state and residue dependence of low-energy CID peptide dissociation patterns. Cleavage N-terminal to proline: analysis of a database of peptide tandem mass spectra. Biomed Environ Mass Spectrom, 16(1-12): p. Contributions of mass spectrometry to peptide and protein structure. Relative specificities of water and ammonia losses from backbone fragments in collision-activated dissociation. High sensitivity collisionally-activated decomposition tandem mass spectrometry on a novel quadrupole/orthogonal-acceleration time-of-flight mass spectrometer. Collisional activation with random noise in ion trap mass spectrometry. The ABC’s (and XYZ’s) of peptide sequencing.
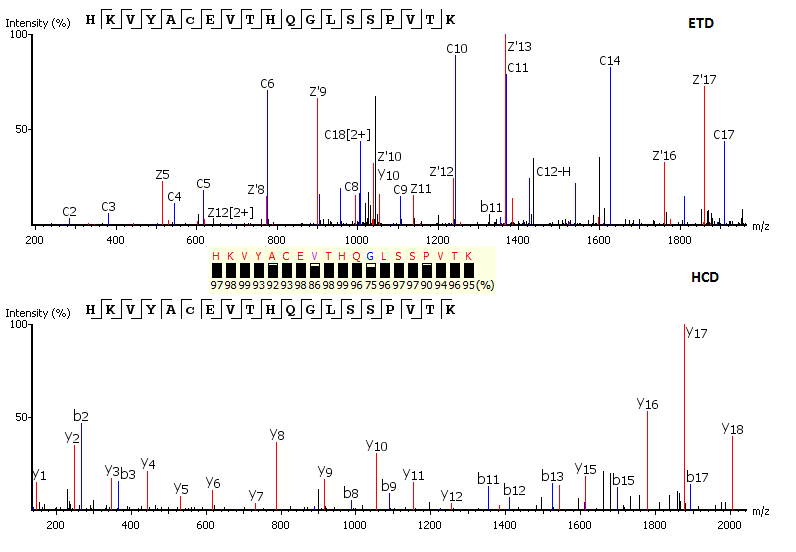
Protein identification using 2D-LC-MS/MS. Determination of dioxins (PCDDs/PCDFs) and PCBs in food and feed using the DR CALUX (R) bioassay: results of an international validation study. The use of a hybrid linear trap/FT-ICR mass spectrometer for on-line high resolution/high mass accuracy bottom-up sequencing. Tandem mass-spectrometry - quadrupole and hybrid instruments. MALDI-TOF mass spectrometry in protein chemistry. An introduction to quadrupole-time-of-flight mass spectrometry. Electrospray interface for liquid chromatographs and mass spectrometers. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Electrospray ionization for mass spectrometry of large biomolecules. Challenges in mass spectrometry-based proteomics. Mass spectrometry-based proteomics: principles, perspectives, and challenges.


 0 kommentar(er)
0 kommentar(er)
